PaccMannRL: Designing anticancer drugs from transcriptomic data via reinforcement learning
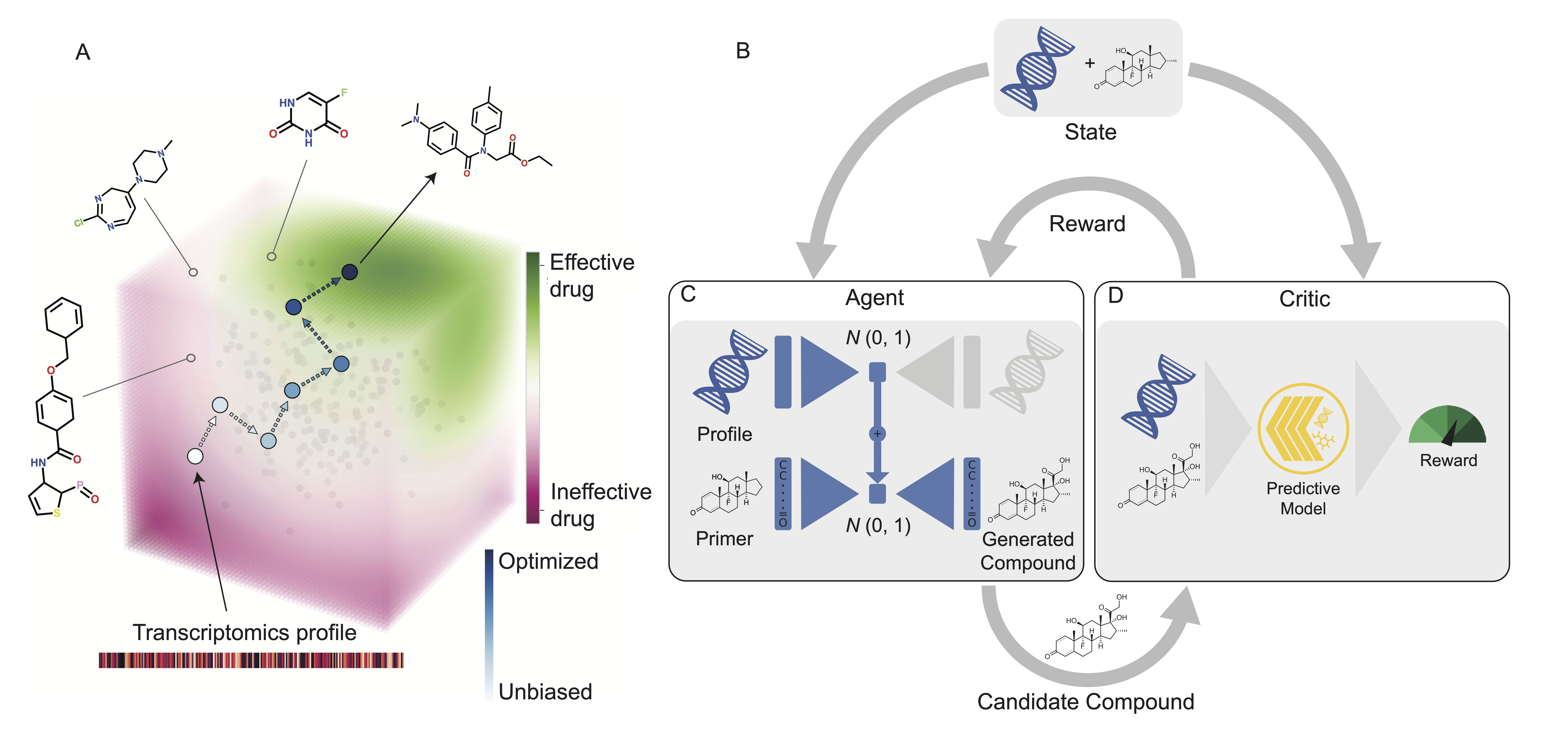
With the advent of deep generative models in computational chemistry, in silico anticancer drug design has undergone an unprecedented transformation. While state-of-the-art deep learning approaches have shown potential in generating compounds with desired chemical properties, they disregard the genetic profile and properties of the target disease. Here, we introduce the first generative model capable of tailoring anticancer compounds for a specific biomolecular profile. Using a RL framework (see figure above), the transcriptomic profiles of cancer cells are used as a context for the generation of candidate molecules. Our molecule generator combines two separately pretrained variational autoencoders (VAEs) - the first VAE encodes transcriptomic profiles into a smooth, latent space which in turn is used to condition a second VAE to generate novel molecular structures on the given transcriptomic profile. The generative process is optimized through PaccMann, a previously developed drug sensitivity prediction model to obtain effective anticancer compounds for the given context (i.e., transcriptomic profile). We demonstrate how the molecule generation can be biased towards compounds with high predicted inhibitory effect against individual cell lines or specific cancer sites. We verify our approach by investigating candidate drugs generated against specific cancer types and find the highest structural similarity to existing compounds with known efficacy against these cancer types. We envision our approach to transform in silico anticancer drug design by leveraging the biomolecular characteristics of the disease in order to increase success rates in lead compound discovery.
For more details, please see the full paper.
Code
The code to reproduce all the results can be found here.
The components adopted are:
- pytoda: implementation of I/O and data-related utilities.
- paccmann_predictor: multimodal drug sensitivity predictor based on our previous paper
- paccmann_omics: generative models for omic data.
- paccmann_chemistry: generative models for chemical data.
- paccmann_generator: multimodal generative models implementing PaccMannRL framework.
PaccMann molecules
We offer the visualization of a selection of molecules generated by PaccMann compared with existing drug-like and/or bioactive compounds at the following links:
- Unbiased molecules, a.k.a., not optimized using PaccMann
- Molecules optmized for breast cancer
- Molecules optmized for prostate cancer
- Molecules optmized for lung cancer
- Molecules optmized for autonomic_ganglia
All the visualizations have been produced using TMAP.